Preprocessing#
Intensity#
RescaleIntensity#
- class torchio.transforms.RescaleIntensity(out_min_max: Tuple[float, float] = (0, 1), percentiles: Tuple[float, float] = (0, 100), masking_method: str | Callable[[Tensor], Tensor] | int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None = None, in_min_max: Tuple[float, float] | None = None, **kwargs)[source]#
Bases:
NormalizationTransformRescale intensity values to a certain range.
- Parameters:
out_min_max – Range \((n_{min}, n_{max})\) of output intensities. If only one value \(d\) is provided, \((n_{min}, n_{max}) = (-d, d)\).
percentiles – Percentile values of the input image that will be mapped to \((n_{min}, n_{max})\). They can be used for contrast stretching, as in this scikit-image example. For example, Isensee et al. use
(0.5, 99.5)in their nn-UNet paper. If only one value \(d\) is provided, \((n_{min}, n_{max}) = (0, d)\).masking_method – See
NormalizationTransform.in_min_max – Range \((m_{min}, m_{max})\) of input intensities that will be mapped to \((n_{min}, n_{max})\). If
None, the minimum and maximum input intensities will be used.**kwargs – See
Transformfor additional keyword arguments.
Example
>>> import torchio as tio >>> ct = tio.ScalarImage('ct_scan.nii.gz') >>> ct_air, ct_bone = -1000, 1000 >>> rescale = tio.RescaleIntensity( ... out_min_max=(-1, 1), in_min_max=(ct_air, ct_bone)) >>> ct_normalized = rescale(ct)
ZNormalization#
- class torchio.transforms.ZNormalization(masking_method: str | Callable[[Tensor], Tensor] | int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None = None, **kwargs)[source]#
Bases:
NormalizationTransformSubtract mean and divide by standard deviation.
- Parameters:
masking_method – See
NormalizationTransform.**kwargs – See
Transformfor additional keyword arguments.
HistogramStandardization#
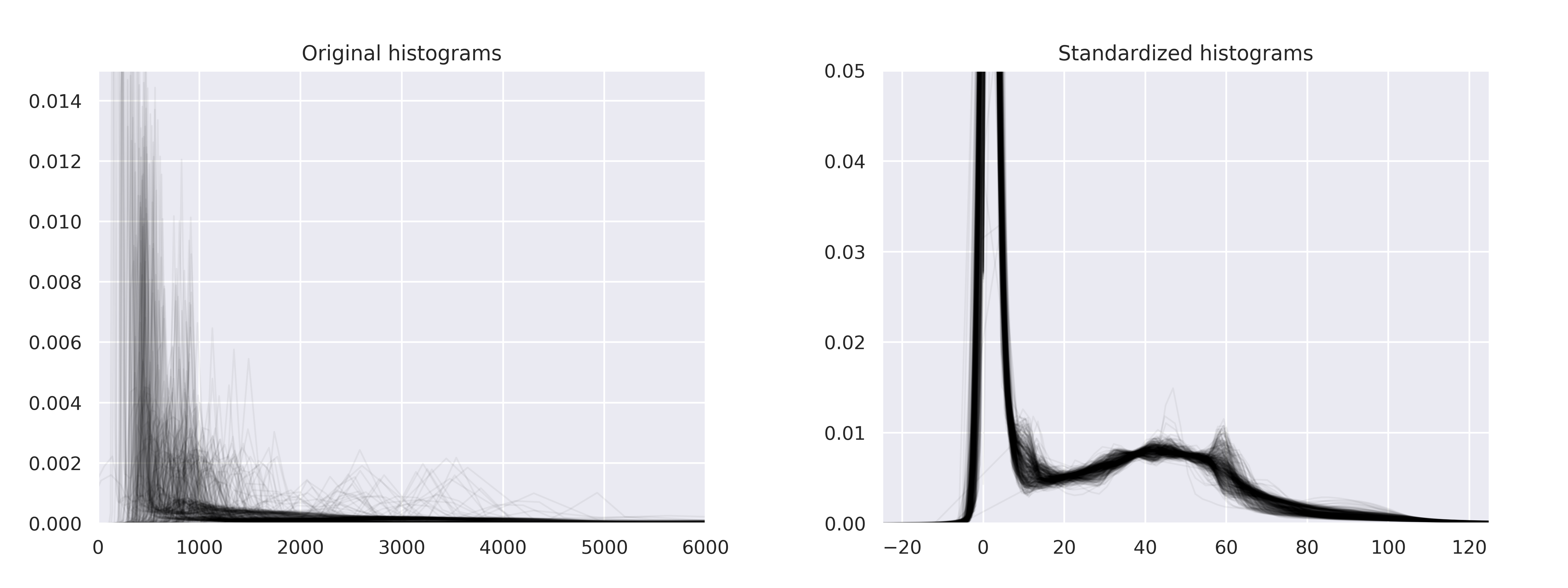
- class torchio.transforms.HistogramStandardization(landmarks: str | Path | Dict[str, str | Path | ndarray], masking_method: str | Callable[[Tensor], Tensor] | int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None = None, **kwargs)[source]#
Bases:
NormalizationTransformPerform histogram standardization of intensity values.
Implementation of New variants of a method of MRI scale standardization.
See example in
torchio.transforms.HistogramStandardization.train().- Parameters:
landmarks – Dictionary (or path to a PyTorch file with
.ptor.pthextension in which a dictionary has been saved) whose keys are image names in the subject and values are NumPy arrays or paths to NumPy arrays defining the landmarks after training withtorchio.transforms.HistogramStandardization.train().masking_method – See
NormalizationTransform.**kwargs – See
Transformfor additional keyword arguments.
Example
>>> import torch >>> import torchio as tio >>> landmarks = { ... 't1': 't1_landmarks.npy', ... 't2': 't2_landmarks.npy', ... } >>> transform = tio.HistogramStandardization(landmarks) >>> torch.save(landmarks, 'path_to_landmarks.pth') >>> transform = tio.HistogramStandardization('path_to_landmarks.pth')
- classmethod train(images_paths: Sequence[str | Path], cutoff: Tuple[float, float] | None = None, mask_path: str | Path | Sequence[str | Path] | None = None, masking_function: Callable | None = None, output_path: str | Path | None = None) ndarray[source]#
Extract average histogram landmarks from images used for training.
- Parameters:
images_paths – List of image paths used to train.
cutoff – Optional minimum and maximum quantile values, respectively, that are used to select a range of intensity of interest. Equivalent to \(pc_1\) and \(pc_2\) in Nyúl and Udupa’s paper.
mask_path – Path (or list of paths) to a binary image that will be used to select the voxels use to compute the stats during histogram training. If
None, all voxels in the image will be used.masking_function – Function used to extract voxels used for histogram training.
output_path – Optional file path with extension
.txtor.npy, where the landmarks will be saved.
Example
>>> import torch >>> import numpy as np >>> from pathlib import Path >>> from torchio.transforms import HistogramStandardization >>> >>> t1_paths = ['subject_a_t1.nii', 'subject_b_t1.nii.gz'] >>> t2_paths = ['subject_a_t2.nii', 'subject_b_t2.nii.gz'] >>> >>> t1_landmarks_path = Path('t1_landmarks.npy') >>> t2_landmarks_path = Path('t2_landmarks.npy') >>> >>> t1_landmarks = ( ... t1_landmarks_path ... if t1_landmarks_path.is_file() ... else HistogramStandardization.train(t1_paths) ... ) >>> np.save(t1_landmarks_path, t1_landmarks) >>> >>> t2_landmarks = ( ... t2_landmarks_path ... if t2_landmarks_path.is_file() ... else HistogramStandardization.train(t2_paths) ... ) >>> np.save(t2_landmarks_path, t2_landmarks) >>> >>> landmarks_dict = { ... 't1': t1_landmarks, ... 't2': t2_landmarks, ... } >>> >>> transform = HistogramStandardization(landmarks_dict)
Mask#
- class torchio.transforms.Mask(masking_method: str | Callable[[Tensor], Tensor] | int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None, outside_value: float = 0, labels: Sequence[int] | None = None, **kwargs)[source]#
Bases:
IntensityTransformSet voxels outside of mask to a constant value.
- Parameters:
masking_method – See
NormalizationTransform.outside_value – Value to set for all voxels outside of the mask.
labels – If a label map is used to generate the mask, sequence of labels to consider. If
None, all values larger than zero will be used for the mask.**kwargs – See
Transformfor additional keyword arguments.
- Raises:
RuntimeWarning – If a 4D image is masked with a 3D mask, the mask will be expanded along the channels (first) dimension, and a warning will be raised.
Example
>>> import torchio as tio >>> subject = tio.datasets.Colin27() >>> subject Colin27(Keys: ('t1', 'head', 'brain'); images: 3) >>> mask = tio.Mask(masking_method='brain') # Use "brain" image to mask >>> transformed = mask(subject) # Set voxels outside of the brain to 0
(
Source code,png)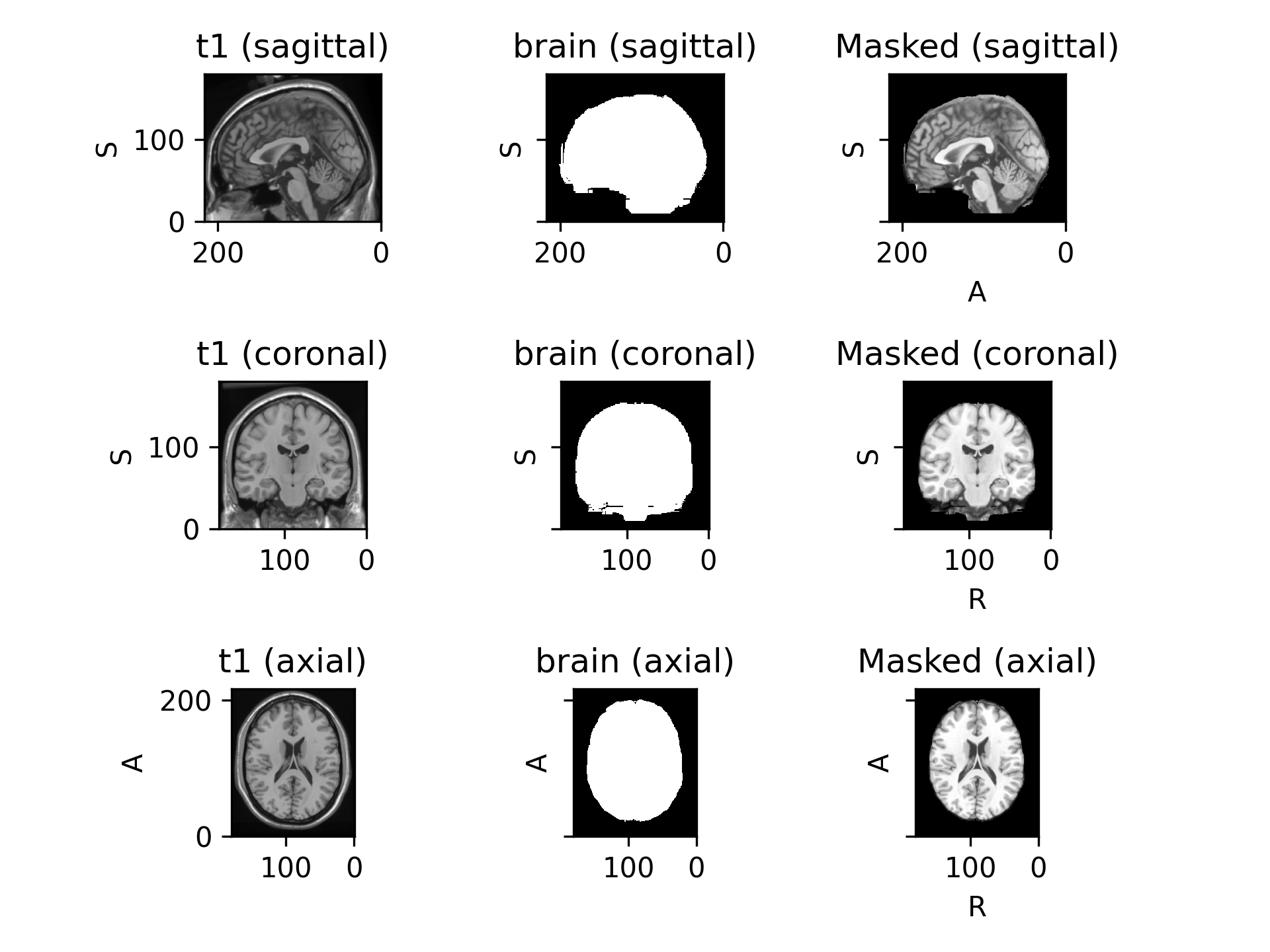
Clamp#
- class torchio.transforms.Clamp(out_min: float | None = None, out_max: float | None = None, **kwargs)[source]#
Bases:
IntensityTransformClamp intensity values into a range \([a, b]\).
For more information, see
torch.clamp().- Parameters:
out_min – Minimum value \(a\) of the output image. If
None, the minimum of the image is used.out_max – Maximum value \(b\) of the output image. If
None, the maximum of the image is used.
Example
>>> import torchio as tio >>> ct = tio.datasets.Slicer('CTChest').CT_chest >>> HOUNSFIELD_AIR, HOUNSFIELD_BONE = -1000, 1000 >>> clamp = tio.Clamp(out_min=HOUNSFIELD_AIR, out_max=HOUNSFIELD_BONE) >>> ct_clamped = clamp(ct)
(
Source code,png)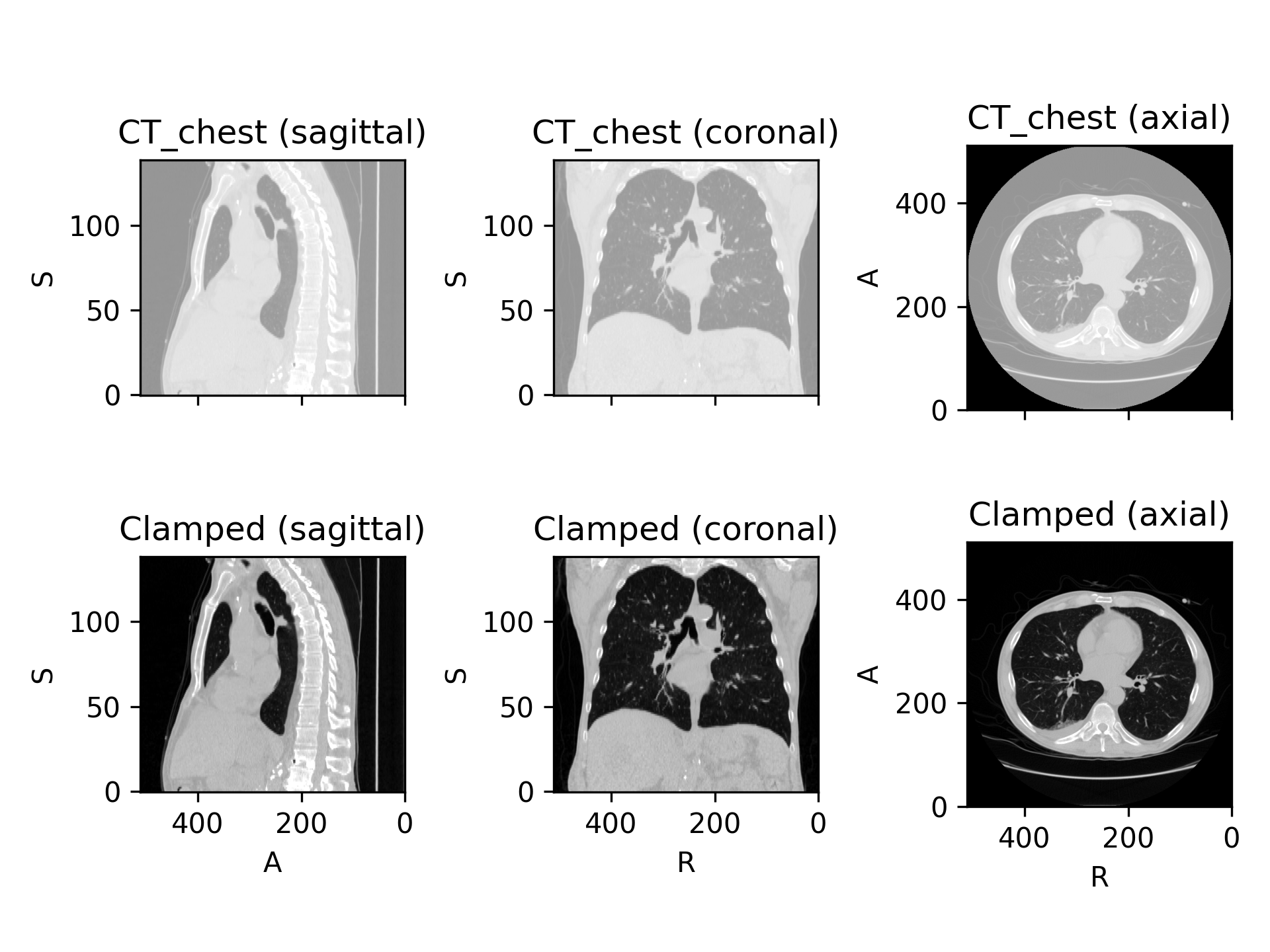
NormalizationTransform#
- class torchio.transforms.preprocessing.intensity.NormalizationTransform(masking_method: str | Callable[[Tensor], Tensor] | int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None = None, **kwargs)[source]#
Bases:
IntensityTransformBase class for intensity preprocessing transforms.
- Parameters:
masking_method –
Defines the mask used to compute the normalization statistics. It can be one of:
None: the mask image is all ones, i.e. all values in the image are used.A string: key to a
torchio.LabelMapin the subject which is used as a mask, OR an anatomical label:'Left','Right','Anterior','Posterior','Inferior','Superior'which specifies a side of the mask volume to be ones.A function: the mask image is computed as a function of the intensity image. The function must receive and return a
torch.Tensor
**kwargs – See
Transformfor additional keyword arguments.
Example
>>> import torchio as tio >>> subject = tio.datasets.Colin27() >>> subject Colin27(Keys: ('t1', 'head', 'brain'); images: 3) >>> transform = tio.ZNormalization() # ZNormalization is a subclass of NormalizationTransform >>> transformed = transform(subject) # use all values to compute mean and std >>> transform = tio.ZNormalization(masking_method='brain') >>> transformed = transform(subject) # use only values within the brain >>> transform = tio.ZNormalization(masking_method=lambda x: x > x.mean()) >>> transformed = transform(subject) # use values above the image mean
Spatial#
CropOrPad#
- class torchio.transforms.CropOrPad(target_shape: int | Tuple[int, int, int] | None = None, padding_mode: str | float = 0, mask_name: str | None = None, labels: Sequence[int] | None = None, **kwargs)[source]#
Bases:
SpatialTransformModify the field of view by cropping or padding to match a target shape.
This transform modifies the affine matrix associated to the volume so that physical positions of the voxels are maintained.
- Parameters:
target_shape – Tuple \((W, H, D)\). If a single value \(N\) is provided, then \(W = H = D = N\). If
None, the shape will be computed from themask_name(and thelabels, iflabelsis notNone).padding_mode – Same as
padding_modeinPad.mask_name – If
None, the centers of the input and output volumes will be the same. If a string is given, the output volume center will be the center of the bounding box of non-zero values in the image namedmask_name.labels – If a label map is used to generate the mask, sequence of labels to consider.
**kwargs – See
Transformfor additional keyword arguments.
Example
>>> import torchio as tio >>> subject = tio.Subject( ... chest_ct=tio.ScalarImage('subject_a_ct.nii.gz'), ... heart_mask=tio.LabelMap('subject_a_heart_seg.nii.gz'), ... ) >>> subject.chest_ct.shape torch.Size([1, 512, 512, 289]) >>> transform = tio.CropOrPad( ... (120, 80, 180), ... mask_name='heart_mask', ... ) >>> transformed = transform(subject) >>> transformed.chest_ct.shape torch.Size([1, 120, 80, 180])
Warning
If
target_shapeisNone, subjects in the dataset will probably have different shapes. This is probably fine if you are using patch-based training. If you are using full volumes for training and a batch size larger than one, an error will be raised by theDataLoaderwhile trying to collate the batches.(
Source code,png)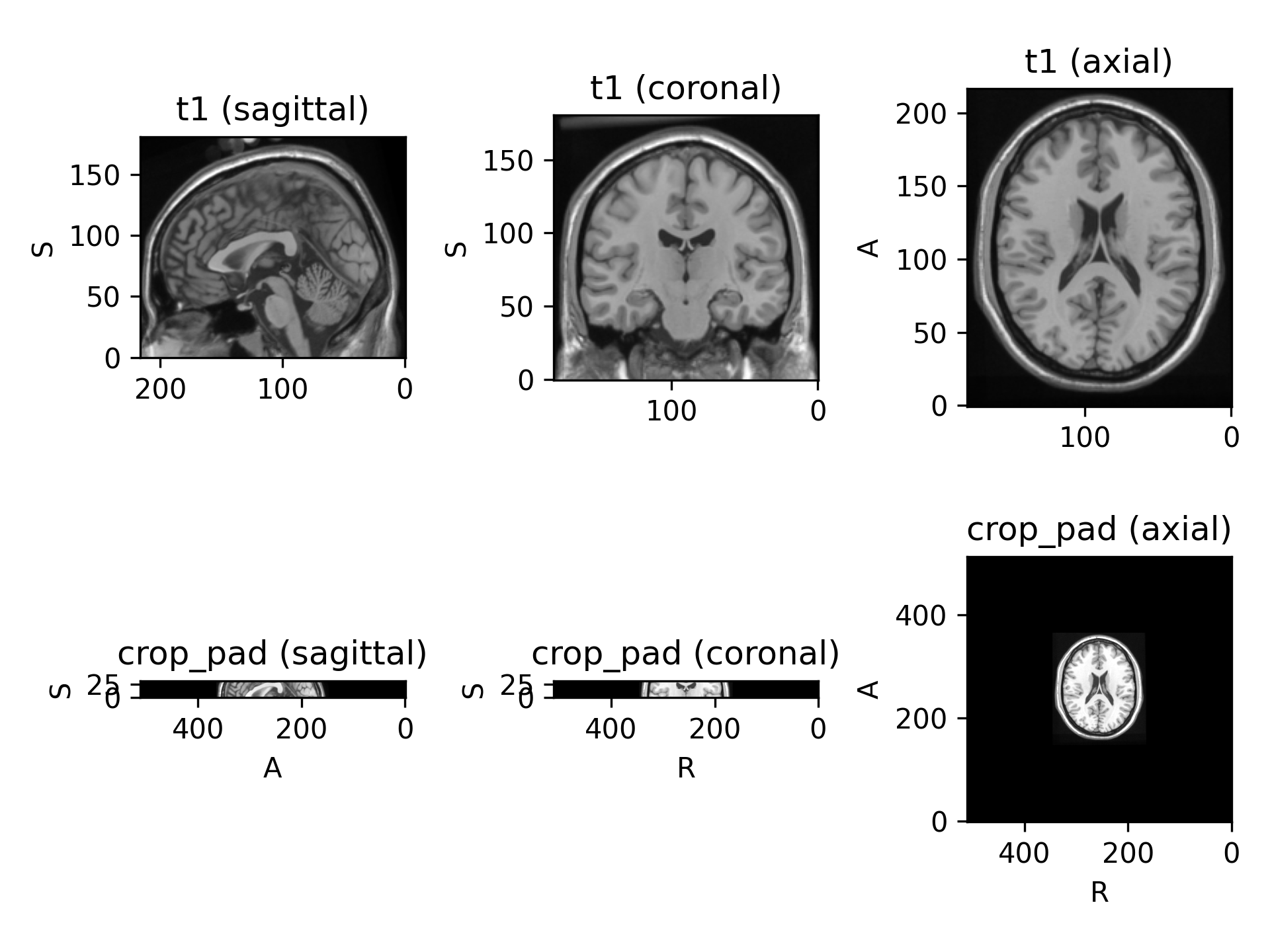
- static _get_six_bounds_parameters(parameters: ndarray) Tuple[int, int, int, int, int, int][source]#
Compute bounds parameters for ITK filters.
- Parameters:
parameters – Tuple \((w, h, d)\) with the number of voxels to be cropped or padded.
- Returns:
Tuple \((w_{ini}, w_{fin}, h_{ini}, h_{fin}, d_{ini}, d_{fin})\), where \(n_{ini} = \left \lceil \frac{n}{2} \right \rceil\) and \(n_{fin} = \left \lfloor \frac{n}{2} \right \rfloor\).
Example
>>> p = np.array((4, 0, 7)) >>> CropOrPad._get_six_bounds_parameters(p) (2, 2, 0, 0, 4, 3)
ToCanonical#
- class torchio.transforms.ToCanonical(p: float = 1, copy: bool = True, include: Sequence[str] | None = None, exclude: Sequence[str] | None = None, keys: Sequence[str] | None = None, keep: Dict[str, str] | None = None, parse_input: bool = True, label_keys: Sequence[str] | None = None)[source]#
Bases:
SpatialTransformReorder the data to be closest to canonical (RAS+) orientation.
This transform reorders the voxels and modifies the affine matrix so that the voxel orientations are nearest to:
First voxel axis goes from left to Right
Second voxel axis goes from posterior to Anterior
Third voxel axis goes from inferior to Superior
See NiBabel docs about image orientation for more information.
- Parameters:
**kwargs – See
Transformfor additional keyword arguments.
Note
The reorientation is performed using
nibabel.as_closest_canonical().
Resample#
- class torchio.transforms.Resample(target: float | Tuple[float, float, float] | str | Path | Image | None = 1, image_interpolation: str = 'linear', label_interpolation: str = 'nearest', pre_affine_name: str | None = None, scalars_only: bool = False, **kwargs)[source]#
Bases:
SpatialTransformResample image to a different physical space.
This is a powerful transform that can be used to change the image shape or spatial metadata, or to apply a spatial transformation.
- Parameters:
target –
Argument to define the output space. Can be one of:
Output spacing \((s_w, s_h, s_d)\), in mm. If only one value \(s\) is specified, then \(s_w = s_h = s_d = s\).
Path to an image that will be used as reference.
Instance of
Image.Name of an image key in the subject.
Tuple
(spatial_shape, affine)defining the output space.
pre_affine_name – Name of the image key (not subject key) storing an affine matrix that will be applied to the image header before resampling. If
None, the image is resampled with an identity transform. See usage in the example below.image_interpolation – See Interpolation.
label_interpolation – See Interpolation.
scalars_only – Apply only to instances of
ScalarImage. Used internally byRandomAnisotropy.**kwargs – See
Transformfor additional keyword arguments.
Example
>>> import torch >>> import torchio as tio >>> transform = tio.Resample(1) # resample all images to 1mm iso >>> transform = tio.Resample((2, 2, 2)) # resample all images to 2mm iso >>> transform = tio.Resample('t1') # resample all images to 't1' image space >>> # Example: using a precomputed transform to MNI space >>> ref_path = tio.datasets.Colin27().t1.path # this image is in the MNI space, so we can use it as reference/target >>> affine_matrix = tio.io.read_matrix('transform_to_mni.txt') # from a NiftyReg registration. Would also work with e.g. .tfm from SimpleITK >>> image = tio.ScalarImage(tensor=torch.rand(1, 256, 256, 180), to_mni=affine_matrix) # 'to_mni' is an arbitrary name >>> transform = tio.Resample(colin.t1.path, pre_affine_name='to_mni') # nearest neighbor interpolation is used for label maps >>> transformed = transform(image) # "image" is now in the MNI space
(
Source code,png)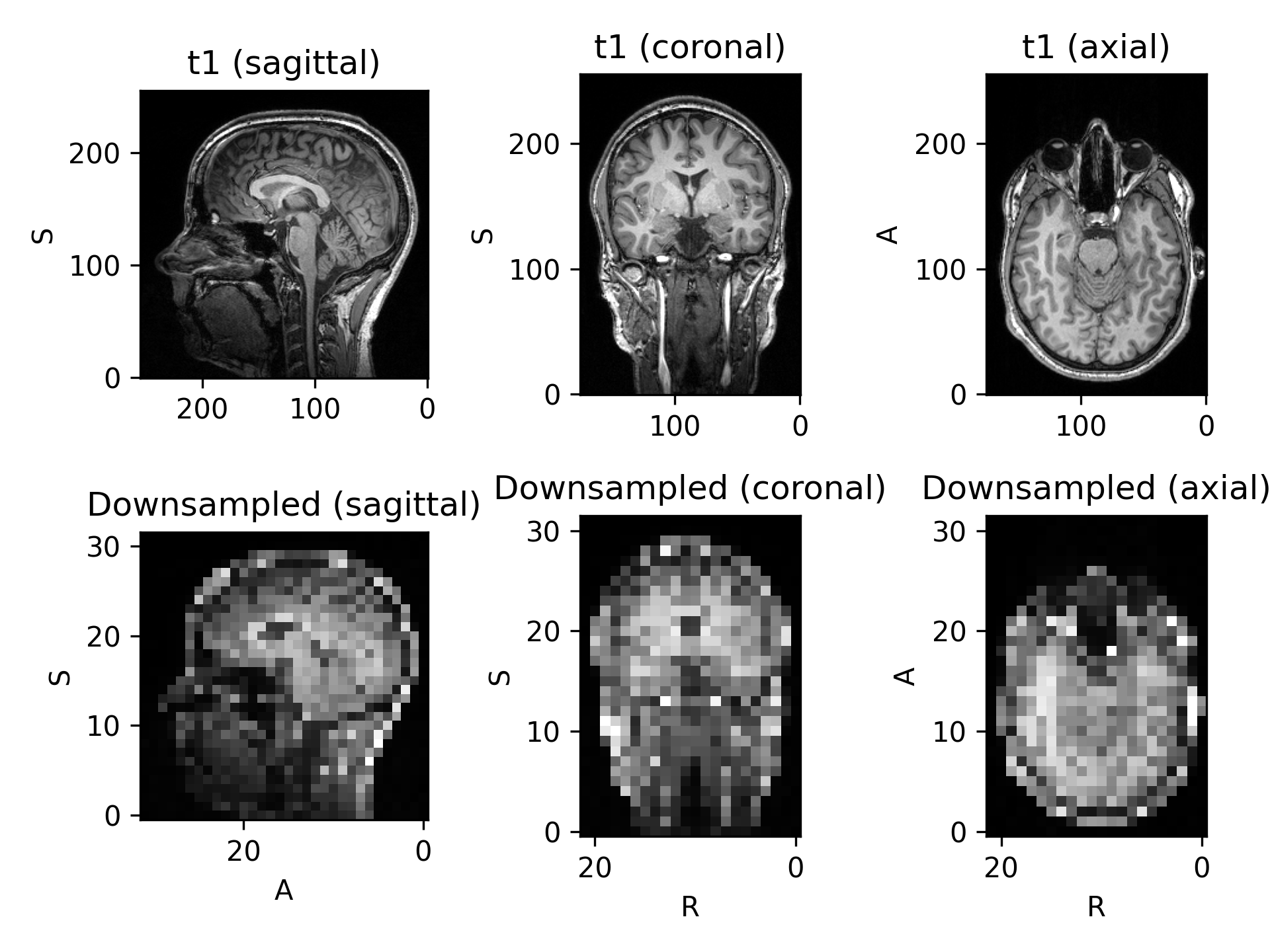
Resize#
- class torchio.transforms.Resize(target_shape: int | Tuple[int, int, int], image_interpolation: str = 'linear', label_interpolation: str = 'nearest', **kwargs)[source]#
Bases:
SpatialTransformResample images so the output shape matches the given target shape.
The field of view remains the same.
Warning
In most medical image applications, this transform should not be used as it will deform the physical object by scaling anistropically along the different dimensions. The solution to change an image size is typically applying
ResampleandCropOrPad.- Parameters:
target_shape – Tuple \((W, H, D)\). If a single value \(N\) is provided, then \(W = H = D = N\). The size of dimensions set to -1 will be kept.
image_interpolation – See Interpolation.
label_interpolation – See Interpolation.
EnsureShapeMultiple#
- class torchio.transforms.EnsureShapeMultiple(target_multiple: int | Tuple[int, int, int], *, method: str = 'pad', **kwargs)[source]#
Bases:
SpatialTransformEnsure that all values in the image shape are divisible by \(n\).
Some convolutional neural network architectures need that the size of the input across all spatial dimensions is a power of \(2\).
For example, the canonical 3D U-Net from Çiçek et al. includes three downsampling (pooling) and upsampling operations:

Pooling operations in PyTorch round down the output size:
>>> import torch >>> x = torch.rand(3, 10, 20, 31) >>> x_down = torch.nn.functional.max_pool3d(x, 2) >>> x_down.shape torch.Size([3, 5, 10, 15])
If we upsample this tensor, the original shape is lost:
>>> x_down_up = torch.nn.functional.interpolate(x_down, scale_factor=2) >>> x_down_up.shape torch.Size([3, 10, 20, 30]) >>> x.shape torch.Size([3, 10, 20, 31])
If we try to concatenate
x_downandx_down_up(to create skip connections), we will get an error. It is therefore good practice to ensure that the size of our images is such that concatenations will be safe.Note
In these examples, it’s assumed that all convolutions in the U-Net use padding so that the output size is the same as the input size.
The image above shows \(3\) downsampling operations, so the input size along all dimensions should be a multiple of \(2^3 = 8\).
Example (assuming
pip install unethas been run before):>>> import torchio as tio >>> import unet >>> net = unet.UNet3D(padding=1) >>> t1 = tio.datasets.Colin27().t1 >>> tensor_bad = t1.data.unsqueeze(0) >>> tensor_bad.shape torch.Size([1, 1, 181, 217, 181]) >>> net(tensor_bad).shape Traceback (most recent call last): File "<stdin>", line 1, in <module> File "/home/fernando/miniconda3/envs/resseg/lib/python3.7/site-packages/torch/nn/modules/module.py", line 727, in _call_impl result = self.forward(*input, **kwargs) File "/home/fernando/miniconda3/envs/resseg/lib/python3.7/site-packages/unet/unet.py", line 122, in forward x = self.decoder(skip_connections, encoding) File "/home/fernando/miniconda3/envs/resseg/lib/python3.7/site-packages/torch/nn/modules/module.py", line 727, in _call_impl result = self.forward(*input, **kwargs) File "/home/fernando/miniconda3/envs/resseg/lib/python3.7/site-packages/unet/decoding.py", line 61, in forward x = decoding_block(skip_connection, x) File "/home/fernando/miniconda3/envs/resseg/lib/python3.7/site-packages/torch/nn/modules/module.py", line 727, in _call_impl result = self.forward(*input, **kwargs) File "/home/fernando/miniconda3/envs/resseg/lib/python3.7/site-packages/unet/decoding.py", line 131, in forward x = torch.cat((skip_connection, x), dim=CHANNELS_DIMENSION) RuntimeError: Sizes of tensors must match except in dimension 1. Got 45 and 44 in dimension 2 (The offending index is 1) >>> num_poolings = 3 >>> fix_shape_unet = tio.EnsureShapeMultiple(2**num_poolings) >>> t1_fixed = fix_shape_unet(t1) >>> tensor_ok = t1_fixed.data.unsqueeze(0) >>> tensor_ok.shape torch.Size([1, 1, 184, 224, 184]) # as expected
- Parameters:
target_multiple – Tuple \((n_w, n_h, n_d)\), so that the size of the output along axis \(i\) is a multiple of \(n_i\). If a single value \(n\) is provided, then \(n_w = n_h = n_d = n\).
method – Either
'crop'or'pad'.**kwargs – See
Transformfor additional keyword arguments.
Example
>>> import torchio as tio >>> image = tio.datasets.Colin27().t1 >>> image.shape (1, 181, 217, 181) >>> transform = tio.EnsureShapeMultiple(8, method='pad') >>> transformed = transform(image) >>> transformed.shape (1, 184, 224, 184) >>> transform = tio.EnsureShapeMultiple(8, method='crop') >>> transformed = transform(image) >>> transformed.shape (1, 176, 216, 176) >>> image_2d = image.data[..., :1] >>> image_2d.shape torch.Size([1, 181, 217, 1]) >>> transformed = transform(image_2d) >>> transformed.shape torch.Size([1, 176, 216, 1])
CopyAffine#
- class torchio.transforms.CopyAffine(target: str, **kwargs)[source]#
Bases:
SpatialTransformCopy the spatial metadata from a reference image in the subject.
Small unexpected differences in spatial metadata across different images of a subject can arise due to rounding errors while converting formats.
If the
shapeandorientationof the images are the same and theiraffineattributes are different but very similar, this transform can be used to avoid errors during safety checks in other transforms and samplers.- Parameters:
target – Name of the image within the subject whose affine matrix will be used.
Example
>>> import torch >>> import torchio as tio >>> import numpy as np >>> np.random.seed(0) >>> affine = np.diag((*(np.random.rand(3) + 0.5), 1)) >>> t1 = tio.ScalarImage(tensor=torch.rand(1, 100, 100, 100), affine=affine) >>> # Let's simulate a loss of precision >>> # (caused for example by NIfTI storing spatial metadata in single precision) >>> bad_affine = affine.astype(np.float16) >>> t2 = tio.ScalarImage(tensor=torch.rand(1, 100, 100, 100), affine=bad_affine) >>> subject = tio.Subject(t1=t1, t2=t2) >>> resample = tio.Resample(0.5) >>> resample(subject).shape # error as images are in different spaces Traceback (most recent call last): File "<stdin>", line 1, in <module> File "/Users/fernando/git/torchio/torchio/data/subject.py", line 101, in shape self.check_consistent_attribute('shape') File "/Users/fernando/git/torchio/torchio/data/subject.py", line 229, in check_consistent_attribute raise RuntimeError(message) RuntimeError: More than one shape found in subject images: {'t1': (1, 210, 244, 221), 't2': (1, 210, 243, 221)} >>> transform = tio.CopyAffine('t1') >>> fixed = transform(subject) >>> resample(fixed).shape (1, 210, 244, 221)
Warning
This transform should be used with caution. Modifying the spatial metadata of an image manually can lead to incorrect processing of the position of anatomical structures. For example, a machine learning algorithm might incorrectly predict that a lesion on the right lung is on the left lung.
Note
For more information, see some related discussions on GitHub:
Crop#
- class torchio.transforms.Crop(cropping: int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None, **kwargs)[source]#
Bases:
BoundsTransformCrop an image.
- Parameters:
cropping – Tuple \((w_{ini}, w_{fin}, h_{ini}, h_{fin}, d_{ini}, d_{fin})\) defining the number of values cropped from the edges of each axis. If the initial shape of the image is \(W \times H \times D\), the final shape will be \((- w_{ini} + W - w_{fin}) \times (- h_{ini} + H - h_{fin}) \times (- d_{ini} + D - d_{fin})\). If only three values \((w, h, d)\) are provided, then \(w_{ini} = w_{fin} = w\), \(h_{ini} = h_{fin} = h\) and \(d_{ini} = d_{fin} = d\). If only one value \(n\) is provided, then \(w_{ini} = w_{fin} = h_{ini} = h_{fin} = d_{ini} = d_{fin} = n\).
**kwargs – See
Transformfor additional keyword arguments.
See also
If you want to pass the output shape instead, please use
CropOrPadinstead.
Pad#
- class torchio.transforms.Pad(padding: int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None, padding_mode: str | float = 0, **kwargs)[source]#
Bases:
BoundsTransformPad an image.
- Parameters:
padding – Tuple \((w_{ini}, w_{fin}, h_{ini}, h_{fin}, d_{ini}, d_{fin})\) defining the number of values padded to the edges of each axis. If the initial shape of the image is \(W \times H \times D\), the final shape will be \((w_{ini} + W + w_{fin}) \times (h_{ini} + H + h_{fin}) \times (d_{ini} + D + d_{fin})\). If only three values \((w, h, d)\) are provided, then \(w_{ini} = w_{fin} = w\), \(h_{ini} = h_{fin} = h\) and \(d_{ini} = d_{fin} = d\). If only one value \(n\) is provided, then \(w_{ini} = w_{fin} = h_{ini} = h_{fin} = d_{ini} = d_{fin} = n\).
padding_mode – See possible modes in NumPy docs. If it is a number, the mode will be set to
'constant'.**kwargs – See
Transformfor additional keyword arguments.
See also
If you want to pass the output shape instead, please use
CropOrPadinstead.
Label#
RemapLabels#
- class torchio.transforms.RemapLabels(remapping: Dict[int, int], masking_method: str | Callable[[Tensor], Tensor] | int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None = None, **kwargs)[source]#
Bases:
LabelTransformModify labels in a label map.
Masking can be used to split the label into two during the inverse transformation.
- Parameters:
remapping – Dictionary that specifies how labels should be remapped. The keys are the old labels, and the corresponding values replace them.
masking_method –
Defines a mask for where the label remapping is applied. It can be one of:
None: the mask image is all ones, i.e. all values in the image are used.A string: key to a
torchio.LabelMapin the subject which is used as a mask, OR an anatomical label:'Left','Right','Anterior','Posterior','Inferior','Superior'which specifies a half of the mask volume to be ones.A function: the mask image is computed as a function of the intensity image. The function must receive and return a 4D
torch.Tensor.
**kwargs – See
Transformfor additional keyword arguments.
(
Source code,png)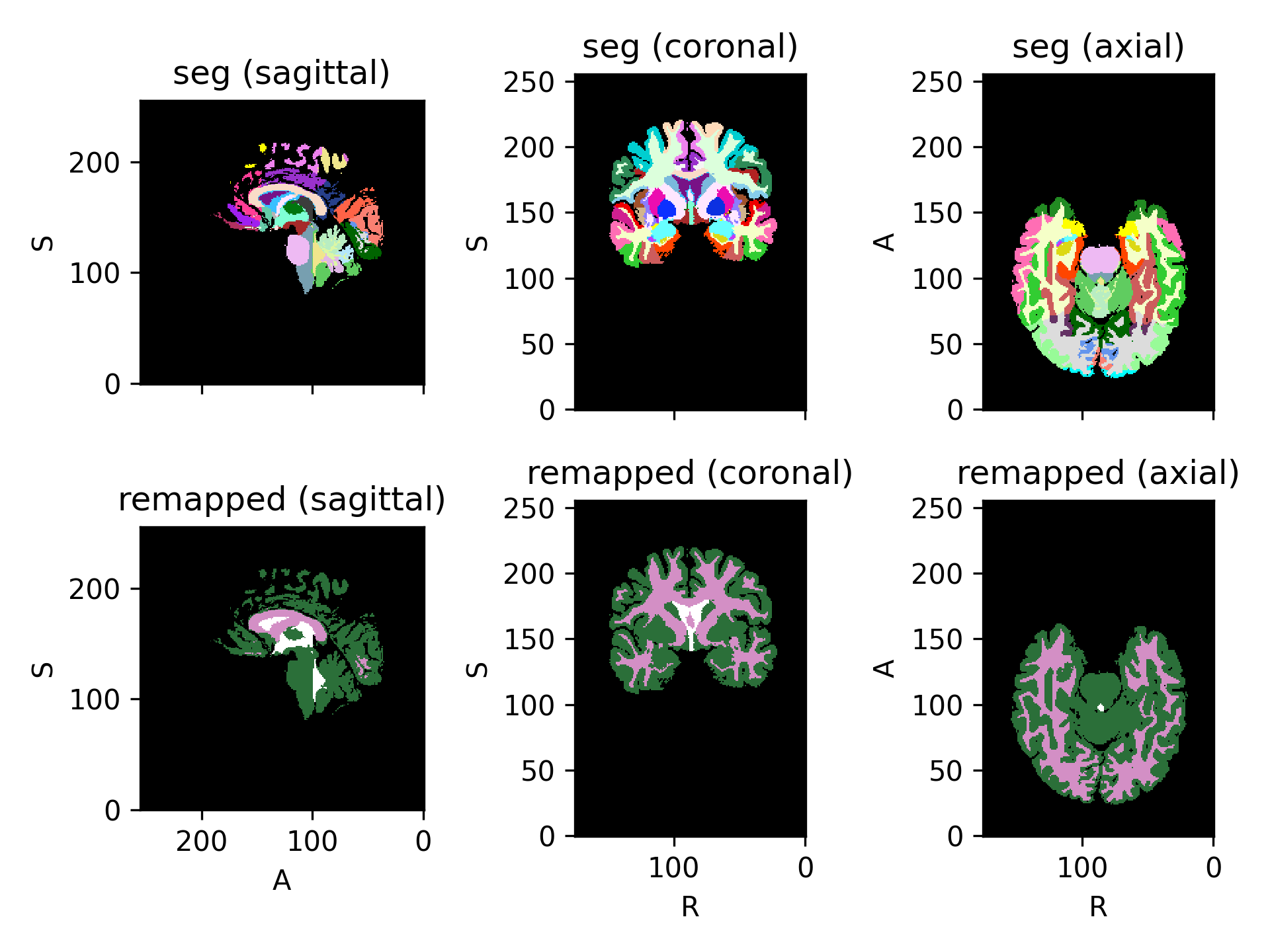
Example
>>> import torch >>> import torchio as tio >>> def get_image(*labels): ... tensor = torch.as_tensor(labels).reshape(1, 1, 1, -1) ... image = tio.LabelMap(tensor=tensor) ... return image ... >>> image = get_image(0, 1, 2, 3, 4) >>> remapping = {1: 2, 2: 1, 3: 1, 4: 7} >>> transform = tio.RemapLabels(remapping) >>> transform(image).data tensor([[[[0, 2, 1, 1, 7]]]])
Warning
The transform will not be correctly inverted if one of the values in
remappingis also in the input image:>>> tensor = torch.as_tensor([0, 1]).reshape(1, 1, 1, -1) >>> subject = tio.Subject(label=tio.LabelMap(tensor=tensor)) >>> mapping = {3: 1} # the value 1 is in the input image >>> transform = tio.RemapLabels(mapping) >>> transformed = transform(subject) >>> back = transformed.apply_inverse_transform() >>> original_label_set = set(subject.label.data.unique().tolist()) >>> back_label_set = set(back.label.data.unique().tolist()) >>> original_label_set {0, 1} >>> back_label_set {0, 3}
Example
>>> import torchio as tio >>> # Target label map has the following labels: >>> # { >>> # 'left_ventricle': 1, 'right_ventricle': 2, >>> # 'left_caudate': 3, 'right_caudate': 4, >>> # 'left_putamen': 5, 'right_putamen': 6, >>> # 'left_thalamus': 7, 'right_thalamus': 8, >>> # } >>> transform = tio.RemapLabels({2:1, 4:3, 6:5, 8:7}) >>> # Merge right side labels with left side labels >>> transformed = transform(subject) >>> # Undesired behavior: The inverse transform will remap ALL left side labels to right side labels >>> # so the label map only has right side labels. >>> inverse_transformed = transformed.apply_inverse_transform() >>> # Here's the *right* way to do it with masking: >>> transform = tio.RemapLabels({2:1, 4:3, 6:5, 8:7}, masking_method="Right") >>> # Remap the labels on the right side only (no difference yet). >>> transformed = transform(subject) >>> # Apply the inverse on the right side only. The labels are correctly split into left/right. >>> inverse_transformed = transformed.apply_inverse_transform()
RemoveLabels#
- class torchio.transforms.RemoveLabels(labels: Sequence[int], background_label: int = 0, masking_method: str | Callable[[Tensor], Tensor] | int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None = None, **kwargs)[source]#
Bases:
RemapLabelsRemove labels from a label map.
The removed labels are remapped to the background label.
This transformation is not invertible.
- Parameters:
labels – A sequence of label integers that will be removed.
background_label – integer that specifies which label is considered to be background (typically,
0).masking_method – See
RemapLabels.**kwargs – See
Transformfor additional keyword arguments.
(
Source code,png)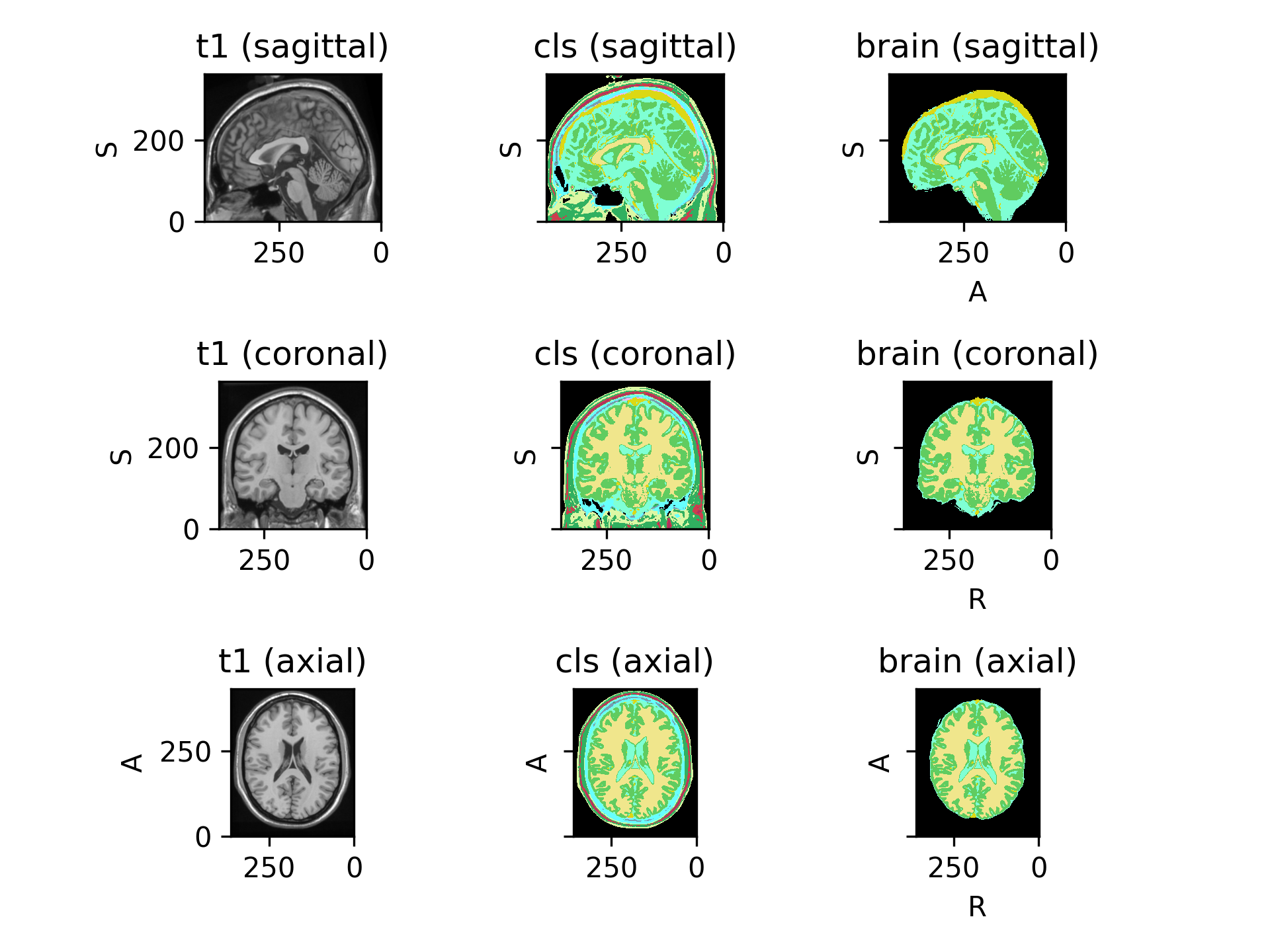
SequentialLabels#
- class torchio.transforms.SequentialLabels(masking_method: str | Callable[[Tensor], Tensor] | int | Tuple[int, int, int] | Tuple[int, int, int, int, int, int] | None = None, **kwargs)[source]#
Bases:
LabelTransformRemap labels in a label map so they become consecutive.
For example, if a label map has labels
(0, 3, 5), then this will apply aRemapLabelstransform withremapping={3: 1, 5: 2}, and therefore the output image will have labels(0, 1, 2).Example
>>> import torch >>> import torchio as tio >>> def get_image(*labels): ... tensor = torch.as_tensor(labels).reshape(1, 1, 1, -1) ... image = tio.LabelMap(tensor=tensor) ... return image ... >>> img_with_bg = get_image(0, 5, 10) >>> transform = tio.SequentialLabels() >>> transform(img_with_bg).data tensor([[[[0, 1, 2]]]]) >>> img_without_bg = get_image(7, 11, 99) >>> transform(img_without_bg).data tensor([[[[0, 1, 2]]]])
Note
This transformation is always fully invertible.
Warning
The background is typically represented with the label
0. There will be zeros in the output image even if they are none in the input.- Parameters:
masking_method – See
RemapLabels.**kwargs – See
Transformfor additional keyword arguments.
OneHot#
Contour#
- class torchio.transforms.Contour(p: float = 1, copy: bool = True, include: Sequence[str] | None = None, exclude: Sequence[str] | None = None, keys: Sequence[str] | None = None, keep: Dict[str, str] | None = None, parse_input: bool = True, label_keys: Sequence[str] | None = None)[source]#
Bases:
LabelTransformKeep only the borders of each connected component in a binary image.
- Parameters:
**kwargs – See
Transformfor additional keyword arguments.
KeepLargestComponent#
- class torchio.transforms.KeepLargestComponent(p: float = 1, copy: bool = True, include: Sequence[str] | None = None, exclude: Sequence[str] | None = None, keys: Sequence[str] | None = None, keep: Dict[str, str] | None = None, parse_input: bool = True, label_keys: Sequence[str] | None = None)[source]#
Bases:
LabelTransformKeep only the largest connected component in a binary label map.
- Parameters:
**kwargs – See
Transformfor additional keyword arguments.
Note
For now, this transform only works for binary images, i.e., label maps with a background and a foreground class. If you are interested in extending this transform, please open a new issue.